
Agenda*



Beverage & Snack BreaksBreaks with complimentary beverages and snacks will be interspersed with the talks. |
Beverages
|
||
Snacks
|

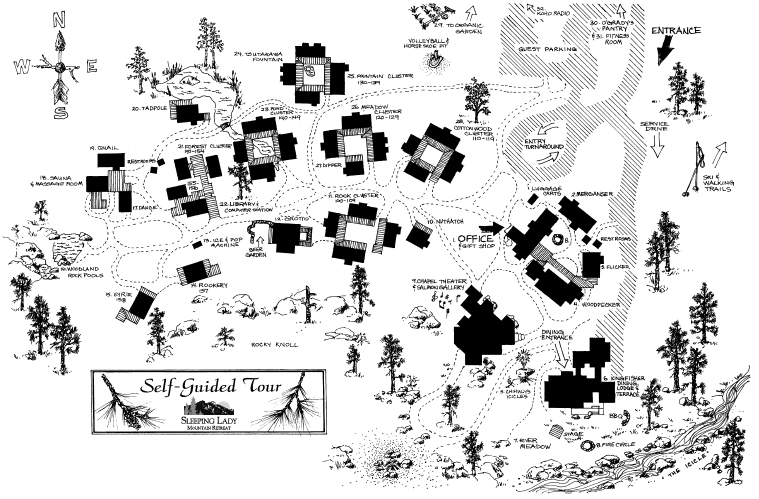
The Rosetta Design Group would like to thank the Participants, the Rosetta Commons Community, the University of Washington, and the Sleeping Lady Mountain Retreat for making this event possible. |
||

The Rosetta Design Group LLC was established in 2007 to bridge the gap between the Rosetta Commons academic community and the pharmaceutical industry.
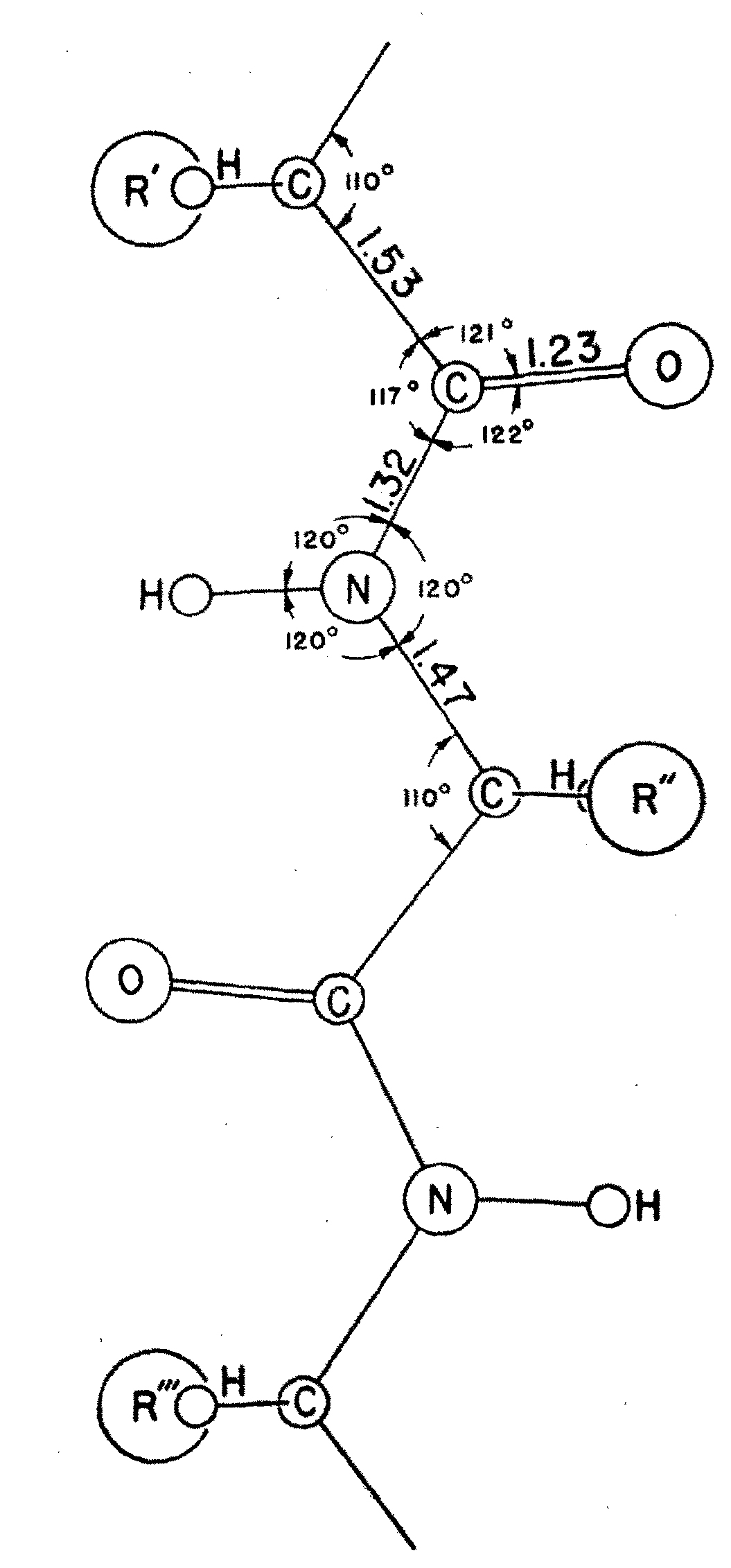
RDG provides services ranging from assistance to computational scientists in protocol development to fully out-sourced R&D in structural modeing and bioinformatics for computational and experimental groups whose demands exceed their in-house capacities. Select examples of areas of R&D: |
|
| Homology modeling | Antibody modeling and epitope mapping |
| Protein design and stabilization | Protein-protein interface design and analysis |
| Protein-peptide interactions, design and analysis | |
Typically, 2-3 days of tailored instruction and hands-on tutorials. Training can be broadcasted live to multiple sites. |
|
Networking and the cutting edge of Rosetta development |
|
Rosetta support resources, events, the Macromolecular Modeling Blog™, and more. |
|


 |
 |
 |
| Andrew Ban Arzeda |
Louis Clark Codexis |
Jacob Corn Genentech |
 |
 |
 |
| Michael Dolan NIAID |
Andrei Golosov Merck |
Katarina Midelfort Pfizer |
 |
 |
|
| Cecile Morales ProteinAI |
Eliud Oloo Sanofi Pasteur |
Arvind Sivasubramanian Adimab |
|
 |
 |
| Vanita Sood EMD Serono |
Jain Tush Adimab |
Alexandre Zanghellini Arzeda |


